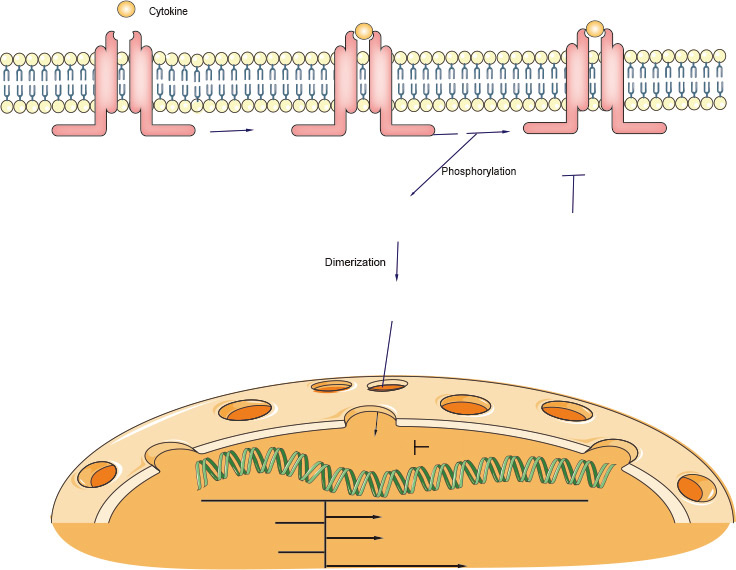
1. JAK-STAT signaling overview
The cytokine-activated Janus kinase (JAK)-signal transducer and activator of transcription (STAT) pathway has an important role in the control of immune responses. Dysregulation of JAK-STAT signaling is associated with various immune disorders. The JAK-STAT signaling pathway transmits information from extracellular chemical signals to the nucleus resulting in DNA transcription and expression of genes involved in immunity, proliferation, differentiation, apoptosis and oncogenesis. This pathway plays a prominent role in mediating signal transduction for many cytokines. Mechanistically, JAK-STAT signaling is relatively simple, the signaling cascade consists of three main components: a JAK binding receptor, a Janus kinase (JAK) and signal transducer and activator of transcription (STAT) protein.
JAK binding receptor
Many cytokines and growth factors transmit signals through the JAK-STAT signaling pathway, including interleukins 2-7 (IL2-7), granulocyte-macrophage colony stimulating factor (GM-CSF), growth hormone (GH), (Epidermal growth factor (EGF), platelet-derived growth factors (PDGF) and interferons (IFN). These cytokines and growth factors have corresponding receptors on the cell membrane. Many cytokines and growth factor cytokine receptors can be divided into two groups: those whose intracellular domains exhibit intrinsic protein tyrosine kinase (PTK) activity, and those whose intracellular domains are devoid of such activity. Many of the latter group of receptors, however, activate intracellular soluble PTKs upon ligand binding.
Janus kinase (JAK)
Janus kinases (JAKs) represent a recently discovered family of PTKs that seem to play a central role in mediating signal transduction of many cytokines, and probably many non-cytokine regulatory molecules. These enzymes harbour two potential active sites and were thus named after Janus, the Roman god with two faces. It is likely that only one of those 'active' sites is functional. Four members of the JAK family have been best characterized to date: JAK1, JAK2, JAK3 and Tyk2. They all exhibit molecular masses in the region of 130 kDa and approximately 40% amino acid sequence homology. They appear to be associated with the cytoplasmic domain of many cytokine receptors, but remain catalytically inactive until binding of the cytokine to the receptor.
JAKs are mainly regulated at the post-translational level through various mechanisms (Figue1). The domains JH1–JH7 are based on sequence similarity of four known JAKs. JH1 is the kinase domain, which contains two tyrosines that can be phosphorylated after ligand stimulation. JH2 is the pseudo-kinase domain. The JH6 and JH7 domains mediate the binding of JAKs to receptors.
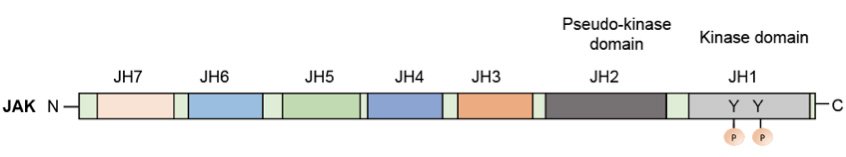 Figue 1. The domain structure of JAKs
Figue 1. The domain structure of JAKs
Signal transducer and activator of transcription (STAT)
STAT stands for 'signal transducers and activators of transcription'. As the name suggests, these proteins (a) form an integral part of cytoplasmic signal transduction initiated by certain regulatory molecules and (b) activate transcription of specific genes in the nucleus. Thus far, at least six distinct mammalian STATs (STAT1–STAT6) have been identified, in size ranging from 84–113 kDa.
All STATs exhibit significant sequence homology and are composed of a number of functional domains (Figure 2). A conserved ('C' or 'con') domain is located at the N-terminus, followed by the DNA-binding domain (D). Y represents a short sequence that contains the tyrosine residue phosphorylated by the Janus kinase. The carboxyl terminus domain (Tr) represents a transcriptional activation domain. The SH2 domain functions to bind phosphotyrosine, thus docking the STAT at the activated receptor surface. An essential tyrosine is located towards the STAT C-terminus (around residue 700), which in turn is then phosplorylated by PTK. Schematic representation of the general domain structure of a STAT protein.
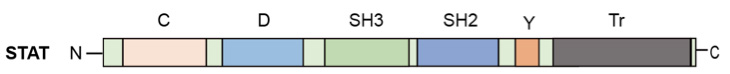 Figure 2. The domain structure of a STAT protein
Figure 2. The domain structure of a STAT protein
2. JAK-STAT signaling cascade
In most instances, ligand binding appears to promote receptor dimerization, bringing their associated JAKs into close proximity. The JAKs then phosphorylate—and hence activate—each other (transphosphorylation). The activated kinases subsequently phosphorylate specific tyrosine residues on the receptor itself. This promotes direct association between one or more members of a family of cytoplasmic proteins (STATs) and the receptor. Once docked at the receptor surface, the STATs are in turn phosphorylated (and hence activated) by the JAKs. As described below, activated STATs then translocate to the nucleus and directly regulate expression of IFN and other cytokine-sensitive genes. Thus the JAK-STAT cascade provides a direct mechanism to translate an extracellular signal into a transcriptional response.
3. Regulation of JAK-STAT signaling
Signaling through the JAK-STAT pathway is tightly controlled by a number of distinct mechanisms. Key regulators of this pathway include suppressors of cytokine signaling (SOCS), Protein inhibitors of activated STATs (PIASs), and protein tyrosine phosphatases (PTPs).
Suppressors of cytokine signaling (SOCS)
SOCS proteins form a classic negative-feedback loop of cytokine signaling. They are typically expressed at low levels and become rapidly induced following cytokine activation of the JAK-STAT pathway. SOCS proteins form a classic negative-feedback loop of cytokine signaling. They are typically expressed at low levels and become rapidly induced following cytokine activation of the JAK-STAT pathway. There are eight members of the CIS–SOCS family (CIS also known as CISH and SOCS1–SOCS7), each of which has a central SH2 domain, an amino (N)-terminal domain of variable length and divergent sequence, and a carboxy (C)-terminal 40-amino-acid module that is known as the SOCS box (Figue 3). Diagram representing the structure of SOCS proteins. At least eight proteins belonging to the SOCS family of proteins are shown. They are characterized by the presence of an SH2 central domain and the SOCS box domain at the C-terminus. A small domain called kinase inhibitory region (KIR), only found in SOCS1 and SOCS3, is shown as a small box at the N-terminal region. SOCS proteins can interact with phosphotyrosine phosphorylated proteins through their SH2 domain and with Elongin BC through their SOCS box domain.
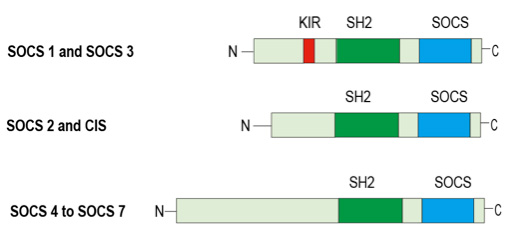 Figue 3. The structure of SOCS proteins
Figue 3. The structure of SOCS proteins
Protein inhibitors of activated STATs (PIASs)
PIAS1 (also known as Gu-binding protein, GBP)82, 83 was identified by yeast two-hybrid assays using STAT1β as the 'bait'. Other members of the PIAS family were subsequently identified through searching the expressed sequence tag (EST) database and complementary DNA screening. The mammalian PIAS family consists of four members: PIAS1, PIAS3, PIASX and PIASY84. Different splicing variants of PIAS proteins exist.
In vivo co-immunoprecipitation studies using antibodies specific for PIAS proteins have identified specific PIAS–STAT interactions in cultured mammalian cells. After cytokine stimulation, PIAS1, PIAS3 and PIASX interact with STAT1, STAT3 and STAT4, respectively.
Protein tyrosine phosphatases (PTPs)
STATs can also be inactivated by PTPs in both the cytoplasm and the nucleus. SHP2 is involved in the dephosphorylation of STAT5 in the cytoplasm.
4. Relationship with diseases
The JAK-STAT pathway is critical for meeting the diverse challenges faced by the immune system, from resisting infection to maintaining immune tolerance, enforcing barrier functions and guarding against cancer.
Nextgeneration sequencing has led to an explosion of genomewide association studies (GWAS) where disease phenotypes are linked to single nucleotide polymorphisms (SNPs) in affected populations. Not surprisingly, GWAS have implicated SNPs within STAT genes in numerous common diseases. Salient examples of this growing list include STAT3 SNPs associated with Crohn's disease, psoriasis and ankylosing spondylitis, STAT4 SNPs associated with rheumatoid arthritis, Crohn's disease, systemic lupus erythematosus and Sjogren's syndrome, and STAT6 SNPs associated with asthma. As with monogenic diseases, SNPs found in JAKs and upstream cytokines/receptors support the central role of STATs in many of these disorders.
5. Therapeutic targeting of JAK-STAT signaling
Given the data implicating JAK-STAT in autoimmune disease and malignancy, it is not surprising that this pathway has become an attractive target for pharmaceuticals. Ruxolitinib, a JAK1 and JAK2 inhibitor with efficacy for the treatment of polycythemia vera and myelofibrosis, was the first FDA approved JAK inhibitor, or Jakinib. The second was Tofacitinib, which blocks JAK3, and to a lesser extent JAK1 and JAK2. First approved for rheumatoid arthritis, it is now in clinical trials for a range of maladies including psoriasis, IBD, transplant rejection, juvenile arthritis and spondyloarthropathy, among others. A third JAK inhibitor, Oclacitinib, is now approved for dermatitis in dogs, and a number of additional Jakinibs, including 'second generation' versions with improved potency and selectivity, are currently under development.
References:
- Gary Walsh. Biopharmaceuticals. 2013.
- Akihiko Yoshimura, Tetsuji Naka and Masato Kubo. SOCS proteins, cytokine signaling and immune regulation. Nature reviews, Immunology, 2017.
- Alejandro V. Villarino, Yuka Kanno and John J.O'Shea. Mechanisms of Jak/STAT signaling in immunity and disease. JImmunol. 2016.
- Ke Shuai and Bin Liu. Regulation of JAK-STAT signaling in the immune system. Nature reviews, 2003.