What is p53?
p53 is a tumor suppressor protein encoded in humans by the TP53 gene.
It had been identified in the 1970s as a binding partner for the SV40 tumor virus oncoprotein large T antigen and an important target for tumor-igenic processes induced by DNA tumor viruses, research in the late 1980s and 1990s validated p53 as a tumor suppressor gene, as p53 mutations were found in up to 50% of human cancers. Mechanistically, p53 acts as a transcription factor that activates and represses a growing number of target genes implicated in cell cycle control. The task of p53 is to control the integrity and correctness of all processes in each individual cell and in the organism as a whole. Information about the state of ongoing events in the cell is gathered through multiple signaling pathways that convey signals modifying activities of p53. The strategy of p53 ensures genetic identity of cells and prevents the selection of abnormal cells. As an anti-cancer promotion agent, it can activate DNA repair proteins when DNA has sustained damage, induce growth arrest by holding the cell cycle at the G1/S regulation point on DNA damage recognition, or initiate apoptosis if DNA damage proves to be irreparable.
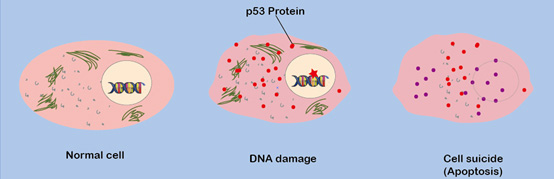 Figure 1. The tumor suppressor protein triggers cell suicide
Figure 1. The tumor suppressor protein triggers cell suicide
p53 family
The p53 family comprises 3 members: p53, p63 and p73. p53 has long been known to be a cellular sensor for DNA damage and has been dubbed the guardian of the genome because of its ability to protect the cell by responding to cellular insults by inducing cell cycle arrest or apoptosis. In 1997, almost 20 years after the discovery of p53, two genes named p63 and p73 were discovered This finding created much excitement, as there are various regions within p63 and p73 that bear significant homology to the well-studied p53. Like p53, p63 and p73 have a transactivation domain, a DNA binding domain, and an oligomerization domain. Both share significant homology with p53 particularly in the DNA binding domain, though p63 and p73 are more homologous to one another than to p53. Because these genes are similar to p53, much speculation grew about their ability to behave like their well-known sibling p53 in their ability to act as tumor suppressor genes. Moreover, these genes were found to reside on chromosomes that are frequently lost or mutated in human cancers. p53 family members play different roles in some of these viruses' life cycles and cellular transformation. p63 and p73 are both much more complex than p53, and it is clear that there is interplay between these family members. Much more investigation is needed to understand this family as a whole in processes such as development and cancer.
p53 signaling pathway
1. p53 signaling cascade
The p53 protein when synthesized in cells has a very short half-life, of 6 to 20 minutes. A wide variety of cellular stress signals appear to activate p53 (as measured by an increased half-life and transcriptional activity) such as UV, Ƴ-irradiation, genotoxic drugs, nutrition deprivation, heat or cold shock, these stress signals are the cause of DNA damage, hypoxia, nitric oxide signaling, and oncogene activation in a cell. In response to UV,gamma irradiation and genotoxic drugs, which make single or double stranded breaks in DNA, the ATM protein kinase CHK2 is activated and in the absence of this kinase p53 activation is delayed or reduced. Similarly after UV irradiation the ATR kinase CHK1 is induced and its absence alters the p53 response.
MDM-2 is the product of a p53-activated gene. The resulting increase of p53 activity leads to upregulation of MDM-2, MDM-2 is a p53 inducible gene whose protein product binds to p53 and acts as an E-3 ubiquitin ligase that adds ubiquitin to p53 and results in its degradation. This produces an autoregulatory loop where p53 results in the synthesis of MDM-2, which in turn degrades p53. This is a “fail-safe” to keep p53 levels in check. MDM-X can oligomerize with MDM-2, which leads to stabilization of MDM-2 and accelerated degradation of MDM-X. Therefore, changes in the proportion of these two proteins can lead to fine regulation of levels and activity of p53. An important player that regulates the activity of MDM-2 is the closely related protein MDM-X. This protein has very similar composition, although unlike MDM-2 it does not have the E3 ubiquitin ligase activity. Another modulator of this process is the p14 or p19 ARF (alternate reading frame) gene. The p19 ARF protein binds to MDM-2 and inhibits its ability to ubiquitinate p53. The transcription of this gene is regulated by a number of oncogene transcription factors including the E2F-1 transcription factor.
2. Pathway regulation
Until quite recently the dominant view was that in normal cells, which are not subjected to various stresses, there is no or minimal activity of p53. The p53 gene is transcribed constitutively, and the mRNA is translated to p53 protein. However, the p53 protein is notoriously unstable, which is due to changeable degradation process that occurs in ubiquitin-dependent and ubiquitin-independent systems of 20S and 26S proteasomes.
Degradation of p53 through the interaction with the ubiquitin ligase MDM-2 remains the most studied and, perhaps, the most important mechanism for regulation of its activity. The interaction of p53 with MDM-2 is subject to a fine regulation by a variety of mechanisms. While some of the mechanisms are connected to regulation of MDM-2, others aim modifications in its target—the p53 itself.
An important player that regulates the activity of MDM-2 is the closely related protein MDM-X. This protein has very similar composition, although unlike MDM-2 it does not have the E3 ubiquitin ligase activity.
An important regulator of the MDM-2-dependent degradation of p53 is p14 ARF, the product of an alternative reading frame of the CDKN2A gene that in addition encodes the CDKs inhibitor p16. ARF is a very basic protein that contains 20% arginine and no lysine residues. Transcription of the ARF gene is subject to positive and negative regulation by complexes that contain transcription factor E2F-1. E2F-1 recognizes a specific set of DNA sequences, which regulate a number of genes that are required for the synthesis of substrate precursors for DNA synthesis and DNA replication.
3. Downstream signaling
About 31 genes have been shown to be regulated by p53 and to have p53 Res(responsive elements) that bind the p53 protein. These genes fall into several categories based upon their functions,which are the cause of growth arrest, apoptosis, inhibition of angiogenesis and DNA repair.
Growth arrest
Several of these proteins mediate cell cycle arrest by p53. The p21 protein binds to Cyclin E–cdk2 and blocks it from phosphorylating the Rb protein which is required for entry into S phase. The 14-3-3 sigma protein binds to Cdc25C, the 14-3-3- Cdc25C complex localizes Cdc25C in the cytoplasm where it cannot act upon Cdc2, the block at this checkpoint occurs through the inhibition of Cdc2–cyclin B complex, while several p53-inducible genes, such as GADD45, BTG2, B99.
Apoptosis
The great majority of p53 responsive genes act in the proapoptotic pathway. Although the role of p53 in apoptosis is undisputed, the mechanism by which p53 induces apoptosis is not fully understood. Studies have suggested that p53-mediated apoptosis involves both transcription-dependent and -independent mechanisms. Much effort has gone into characterizing the transcriptional activation of its targets and a number of genes including BAX, Bcl2, BID, NOXA, p53AIP1, PIGS and PUMA have been implicated in p53-mediated apoptosis. Cytochrome c release from mitochondria binds to apaf-1 resulting in apaf-1 cleavage and activation. This in turn cleaves caspase-9 which cleaves caspase-3 leading irreversibly to apoptosis. Thus, the p53 proapoptotic pathway acts upon many fronts that collectively execute programmed cell death.
Inhibition of angiogenesis
p53-regulated genes include an angiogenesis inhibitor thrombospondin (TSP-1), GDAIF, BAI1, inhibitors of tumor invasion and metastasis KAI1, Maspin, an inhibitor of the plasminogen activator protein PAI, and several secreting protein factors which inhibit proliferation of exposed cells.
DNA repair
The genes encoding components of the global genome repair, the p48XPE, and XPC are among the targets of p53. Another set of p53-regulated genes, for example, GADD45 and p53R2 (a ribonucleotide reductase subunit), result in enhanced DNA repair in the cell or in the culture media. They are important for maintenance of the nucleotide pools during the DNA repair.
4. Functions of the p53 pathway
p53 has many mechanisms of anticancer function and plays a role in apoptosis, monitor DNA replication, cell division, and inhibition of angiogenesis. As an anti-cancer promotion agent, it can activate DNA repair proteins when DNA has sustained damage, induce growth arrest by holding the cell cycle at the G1/S regulation point on DNA damage recognition, or initiate apoptosis if DNA damage proves to be irreparable.
References:
- William G Kaelin Jr. The p53 gene family[J]. Oncogene, 1999,18: 7701-7705.
- P. M. Chumakov. Versatile Functions of p53 Protein in Multicellular Organisms[J]. Biochemistry (Mosc), 2007, 72(13): 1399–1421.
- Jean Benard, Setha Douc-Rasy. TP53 Family Members and Human Cancers[J]. Human mutation,2003, 21:182-191.
- Gerard P. Zambetti. The p53 tumor suppressor pathway and cancer.
- Gen Sheng Wu.The p53 tumor suppressor pathway and cancer[J]. Cancer Biology & Therapy, 2004, 3:2,156-161.