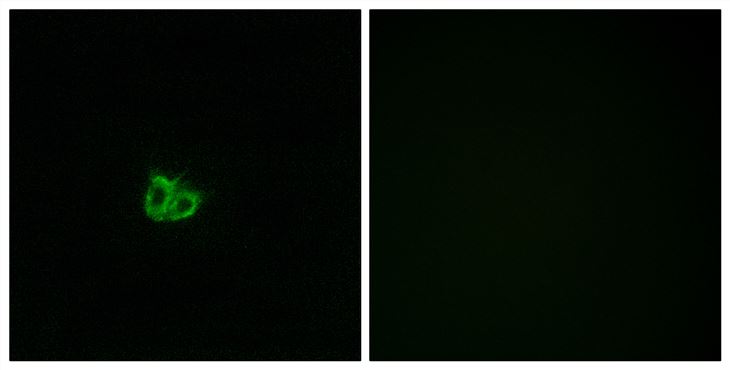
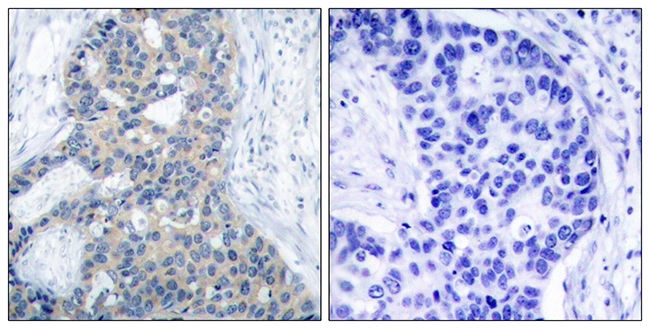
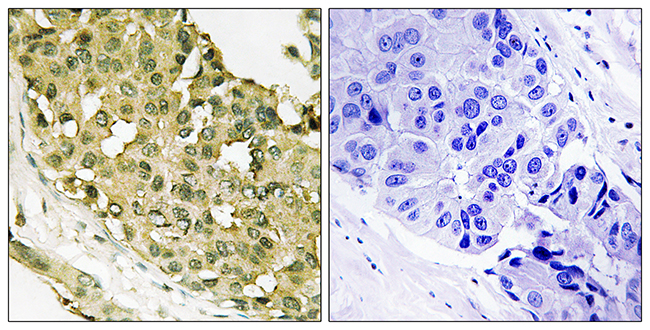
Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity
PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA
Authors: Saal, Lao H.; Johansson, Peter; Holm, Karolina; Gruvberger-Saal, Sofia K.; She, Qing-Bai; Maurer, Matthew; Koujak, Susan; Ferrando, Adolfo A.; Malmstrom, Per; Memeo, Lorenzo; Isola, Jorma; Bendahl, Par-Ola; Rosen, Neal; Hibshoosh, Hanina; Ringner, Markus; Borg, Ake; Parsons, Ramon
Abstract
Pathway-specific therapy is the future of cancer management. The oncogenic phosphatidylinositol 3-kinase (PI3K) pathway is frequently activated in solid tumors; however, currently, no reliable test for PI3K pathway activation exists for human tumors. Taking advantage of the observation that loss of PTEN, the negative regulator of PI3K, results in robust activation of this pathway, we developed and validated a microarray gene expression signature for immunohistochemistry (IHC)-detectable PTEN loss in breast cancer (IBC). The most significant signature gene was PTEN itself, indicating that PTEN mRNA levels are the primary determinant of PTEN protein levels in BC. Some PTEN IHC-positive BCs exhibited the signature of PTEN loss, which was associated to moderately reduced PTEN mRNA levels cooperating with specific types of PIK3CA mutations and/or amplification of HER2. This demonstrates that the signature is more sensitive than PTEN IHC for identifying tumors with pathway activation. In independent data sets of breast, prostate, and bladder carcinoma, prediction of pathway activity by the signature correlated significantly to poor patient outcome. Stathmin, encoded by the signature gene STMN1, was an accurate IHC marker of the signature and had prognostic significance in BC. Stathmin was also pathway-pharmacodynamic in vitro and in vivo. Thus, the signature or its components such as stathmin may be clinically useful tests for stratification of patients for anti-PI3K pathway therapy and monitoring therapeutic efficacy. This study indicates that aberrant PI3K pathway signaling is strongly associated with metastasis and poor survival across carcinoma types, highlighting the enormous potential impact on patient survival that pathway inhibition could achieve.
Identification of potential biomarkers in cervical cancer with combined public mRNA and miRNA expression microarray data analysis
ONCOLOGY LETTERS
Authors: Wang, Sizhe; Chen, Xiaojin
Abstract
Cervical cancer is the fourth most prevalent malignancy in females worldwide. Early diagnosis is key to improving survival rates. Molecular biomarkers are an important method for diagnosing a number of types of cancer, including cervical cancer. The present study utilized public data from three mRNA microarray datasets and one microRNA dataset to analyze the key genes involved in cervical cancer. The mRNA and microRNA expression profile datasets (GSE9750, GSE46857, GSE67522 and GSE30656) were downloaded from the Gene Expression Omnibus database (GEO). Differentially expressed genes (DEGs) and microRNAs (DEMs) were screened using the online tool GEO2R. By using the DEGs consistent across the three mRNA datasets, a functional and pathway enrichment analysis was performed using the Database for Annotation, Visualization and Integrated Discovery. A protein-protein interaction (PPI) network was constructed and module analysis performed using the Search Tool for the Retrieval of Interacting Genes. Validated target genes of the DEMs were identified using the miRecords website. Using the identified target genes of the DEMs, a survival analysis was performed using the OncoLnc online tool. A total of 73 DEGs and 19 DEMs were screened from the microarray expression profile datasets. Integrin-mediated', proteolysis' and phosphoinositide 3 kinase-protein kinase 3' signaling pathways were the most enriched in the DEGs. Three of the DEGs, including Ras homolog family member B (RhoB), stathmin 1 (STMN1) and cyclin D1 (CCNB1) were validated DEM target genes. The OncoLnc survival analysis identified that RhoB was associated with a significantly longer overall survival, whereas STMN1 was associated with a significantly reduced overall survival time in patients with cervical cancer. Finally, data from The Cancer Genome Atlas revealed an association between the mRNA expression levels of RhoB and STMN1, and the overall survival time for patients with cervical cancer. In conclusion, RhoB and STMN1 were identified as key genes that may provide potential targets for cervical cancer diagnosis and treatment.
![]()